Add analytes
It’s possible to upload a structure (EICs will be created automatically) either to individual injections or to many injections at a time in a Batch.
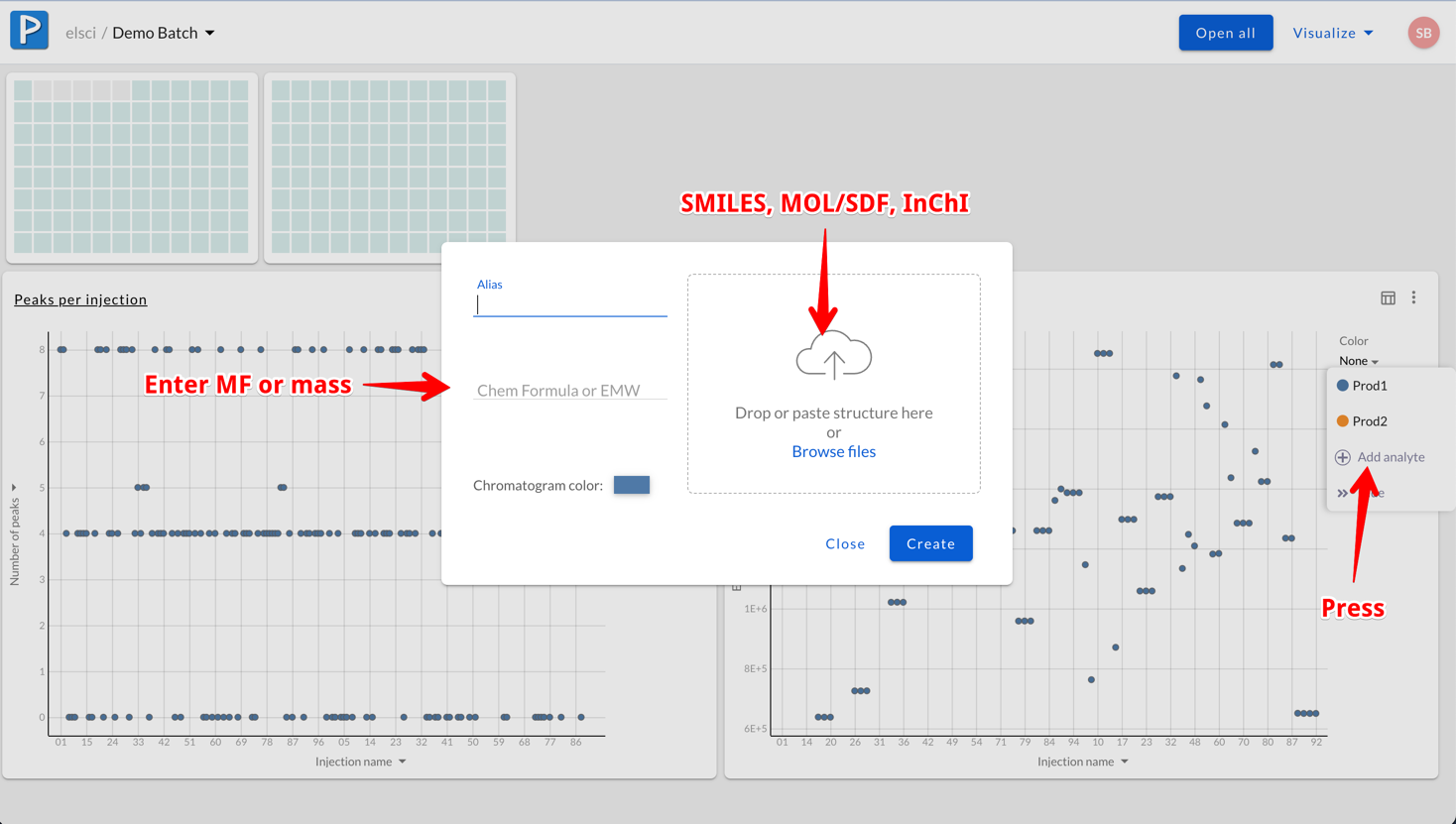
Manually map analytes to wells (few analytes)
When adding the same analyte to many wells, you can manually select which analytes to which wells. Just open the dialog, enter chemical information and then select the wells:
Upload a file with analytes (many analytes)
When adding many analytes, you can upload a file containing structures and their corresponding wells.
Columns/Attributes
The file needs to have 3 attributes or columns:
-
Chemical structure (SMILES, MOL/SDF, InChi, MF aka Molecular Formula, EMW aka Exact Molecular Weight)
-
Alias (it’s more of a role, not a chemical name!). Suppose you analyze 10 crude reactions with different products. If each product has its own name, in the reports you’ll have 10 different columns
Product1 Area,Product2 Area, etc. That’s not what you want! You want them all to have the same logical name likeProduct, then in the report there’ll be just one columnProduct Area, and you can have a separate columnProduct MFto identify which particular formula we’re dealing with. -
Injection - the name of the injection this structure has to go to
File formats (Excel, SSV/TSV)
You can use different formats to upload:
-
SSV (semicolon-separated values) or TSV (tab-separated values) - it’s basically a CSV, but separated with
;or\t -
Excel with the respective columns
-
SDF (both V2000 and V3000) - when using it, the attributes must be called
name(for alias) andinjection(for injection name)
You can either upload/drag’n’drop files or just copy-paste their content directly into Peaksel dialog.
When uploading CSV or Excel, Peaksel will try to automatically determine the chemical format by value in the 1st row and 1st column. But sometimes it’s impossible to tell SMILES apart from MF. If you want to be sure, you can name the column mf, smiles, inchi to help Peaksel.
|
Identifying unknowns
If you use Mass Spec, Peaksel can help you figure out what the suspicious m/z in the spectra correspond to. See Identifying unknown analytes.