Parsing Structures from Waters RAW
It's possible to embed molecular structures (SMILES, InChI) or
molecular formulae (MF) in Waters RAW files. Peaksel can read
those structures and create EIC's based on this information. For
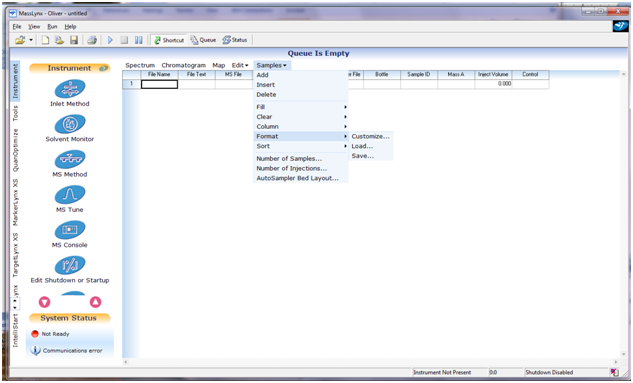
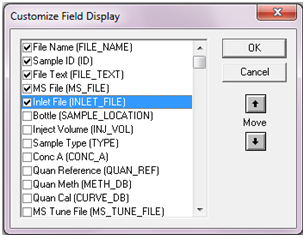
this to work, when creating MassLynx Sample List you need to add
columns
Spare1 (will hold structures) and
Spare2 (will hold substance aliases). Adding columns
is described in the
official MassLynx
documentation
:
The cells in these columns may contain more than 1 Structure/MF
separated with underscores "_". Same goes for
aliases. Example (spreadsheet
):
| Sample Name | Spare1 | Spare2 |
|---|---|---|
| 01-MF | C6O6H12_C2H6O | Glucose_Ethanol |
| 02-SMILES | OC[C@H]1OC(O)[C@H](O)[C@@H](O)[C@@H]1O_OCC | Glucose_Ethanol |
| 02-InChI | InChI=1S/C6H12O6/c7-1-2-3(8)4(9)5(10)6(11)12-2/h2-11H,1H2/t2-,3-,4+,5-,6?/m1/s1_InChI=1S/C2H6O/c1-2-3/h3H,2H2,1H3 | Glucose_Ethanol |


